Research
Highlighted
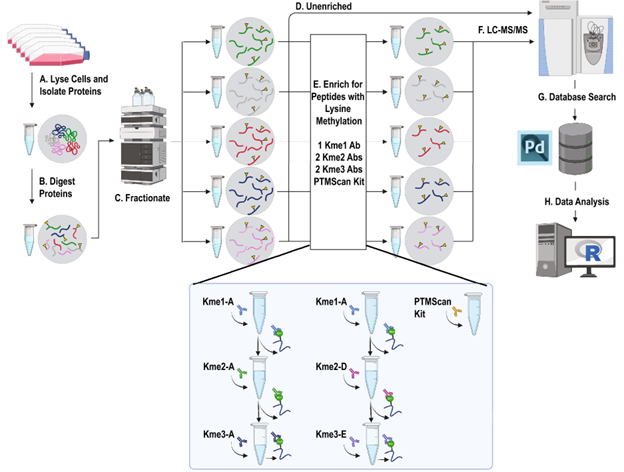
Global lysine methylome profiling using systematically characterized affinity reagents
Scientific Reports
·
07 Jan 2023
·
doi:10.1038/s41598-022-27175-x
All
2025
Quantitative Analysis of Nonhistone Lysine Methylation Sites and Lysine Demethylases in Breast Cancer Cell Lines
Journal of Proteome Research
·
08 Jan 2025
·
doi:10.1021/acs.jproteome.4c00685
2024
Select EZH2 inhibitors enhance viral mimicry effects of DNMT inhibition through a mechanism involving NFAT:AP-1 signaling
Science Advances
·
29 Mar 2024
·
doi:10.1126/sciadv.adk4423
2023
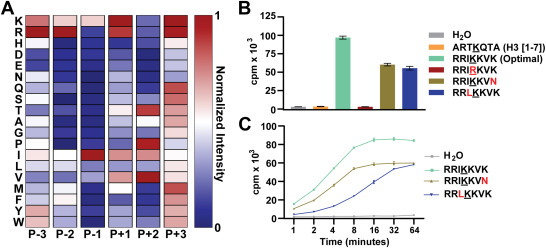
Identification of nonhistone substrates of the lysine methyltransferase PRDM9
Journal of Biological Chemistry
·
01 May 2023
·
doi:10.1016/j.jbc.2023.104651

Global lysine methylome profiling using systematically characterized affinity reagents
Scientific Reports
·
07 Jan 2023
·
doi:10.1038/s41598-022-27175-x
2022
Structural and genome-wide analyses suggest that transposon-derived protein SETMAR alters transcription and splicing
Journal of Biological Chemistry
·
01 May 2022
·
doi:10.1016/j.jbc.2022.101894
2020
A physical basis for quantitative ChIP-sequencing
Journal of Biological Chemistry
·
01 Nov 2020
·
doi:10.1074/jbc.RA120.015353
Click Chemistry-Based Two-Component System for Efficient Inhibition of Human Immunodeficiency Virus (HIV) Reverse Transcriptase (RT)
ACS Omega
·
21 Feb 2020
·
doi:10.1021/acsomega.9b03942
2019
A Read/Write Mechanism Connects p300 Bromodomain Function to H2A.Z Acetylation
iScience
·
01 Nov 2019
·
doi:10.1016/j.isci.2019.10.053
Lysine Methylation Regulators Moonlighting outside the Epigenome
Molecular Cell
·
01 Sep 2019
·
doi:10.1016/j.molcel.2019.08.026
Selective binding of the PHD6 finger of MLL4 to histone H4K16ac links MLL4 and MOF
Nature Communications
·
24 May 2019
·
doi:10.1038/s41467-019-10324-8
2018
A DNA methylation reader complex that enhances gene transcription
Science
·
07 Dec 2018
·
doi:10.1126/science.aar7854
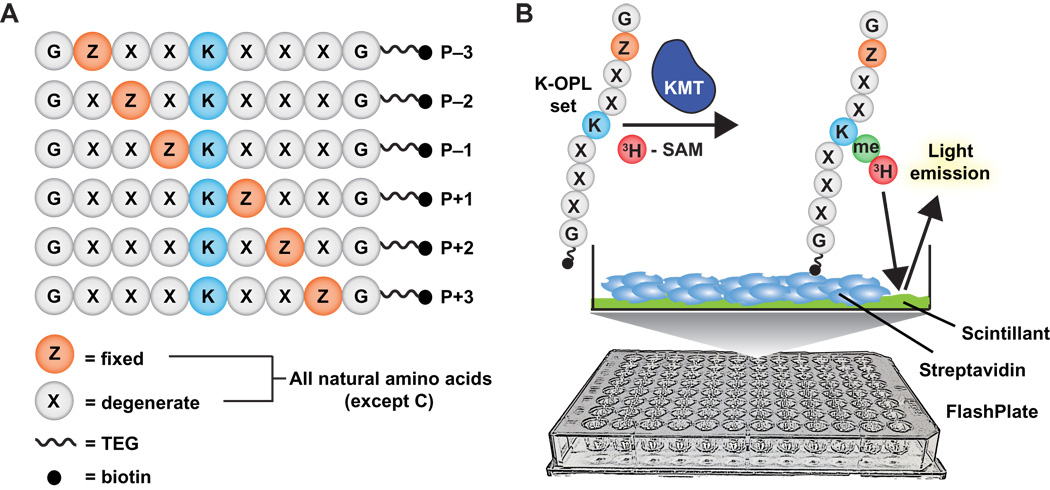
A functional proteomics platform to reveal the sequence determinants of lysine methyltransferase substrate selectivity
Science Advances
·
02 Nov 2018
·
doi:10.1126/sciadv.aav2623
Examining the Roles of H3K4 Methylation States with Systematically Characterized Antibodies
Molecular Cell
·
01 Oct 2018
·
doi:10.1016/j.molcel.2018.08.015
Chromatin structure and its chemical modifications regulate the ubiquitin ligase substrate selectivity of UHRF1
Proceedings of the National Academy of Sciences
·
13 Aug 2018
·
doi:10.1073/pnas.1806373115
Comparative biochemical analysis of UHRF proteins reveals molecular mechanisms that uncouple UHRF2 from DNA methylation maintenance
Nucleic Acids Research
·
28 Feb 2018
·
doi:10.1093/nar/gky151
2017
Analysis of Histone Antibody Specificity with Peptide Microarrays
Journal of Visualized Experiments
·
01 Aug 2017
·
doi:10.3791/55912
2016
Systematic comparison of monoclonal versus polyclonal antibodies for mapping histone modifications by ChIP-seq
Epigenetics & Chromatin
·
04 Nov 2016
·
doi:10.1186/s13072-016-0100-6
Hemi-methylated DNA regulates DNA methylation inheritance through allosteric activation of H3 ubiquitylation by UHRF1
eLife
·
06 Sep 2016
·
doi:10.7554/eLife.17101
Multivalent Chromatin Engagement and Inter-domain Crosstalk Regulate MORC3 ATPase
Cell Reports
·
01 Sep 2016
·
doi:10.1016/j.celrep.2016.08.050
Substrate Specificity Profiling of Histone-Modifying Enzymes by Peptide Microarray
Methods in Enzymology
·
01 Jan 2016
·
doi:10.1016/bs.mie.2016.01.008
ArrayNinja
Methods in Enzymology
·
01 Jan 2016
·
doi:10.1016/bs.mie.2016.02.002
2015
Nuclease-containing media for resettable operation of DNA logic gates
Chemical Communications
·
01 Jan 2015
·
doi:10.1039/c4cc09283j
2013
Deoxyribozyme Cascade for Visual Detection of Bacterial RNA
ChemBioChem
·
17 Sep 2013
·
doi:10.1002/cbic.201300471
Two-component covalent inhibitor
Bioorganic & Medicinal Chemistry
·
01 Apr 2013
·
doi:10.1016/j.bmc.2013.01.021
Operating Cooperatively (OC) Sensor for Highly Specific Recognition of Nucleic Acids
PLoS ONE
·
18 Feb 2013
·
doi:10.1371/journal.pone.0055919
SNP Analysis Using a Molecular Beacon-Based Operating Cooperatively (OC) Sensor
Methods in Molecular Biology
·
01 Jan 2013
·
doi:10.1007/978-1-62703-535-4_6
2012
Molecular Logic Gates for DNA Analysis: Detection of Rifampin Resistance in M. tuberculosis DNA
Angewandte Chemie International Edition
·
09 Aug 2012
·
doi:10.1002/anie.201203708
2010
RNA‐Cleaving Deoxyribozyme Sensor for Nucleic Acid Analysis: The Limit of Detection
ChemBioChem
·
01 Apr 2010
·
doi:10.1002/cbic.201000006
